The Epigenetic Plasticity lab, led by Dr Melanie Eckersley-Maslin, aims to understand how cells adapt and respond to change. We use insights from developmental biology to give a different perspective into cancer biology, ultimately uncovering new molecules or pathways that could be exploited therapeutically.
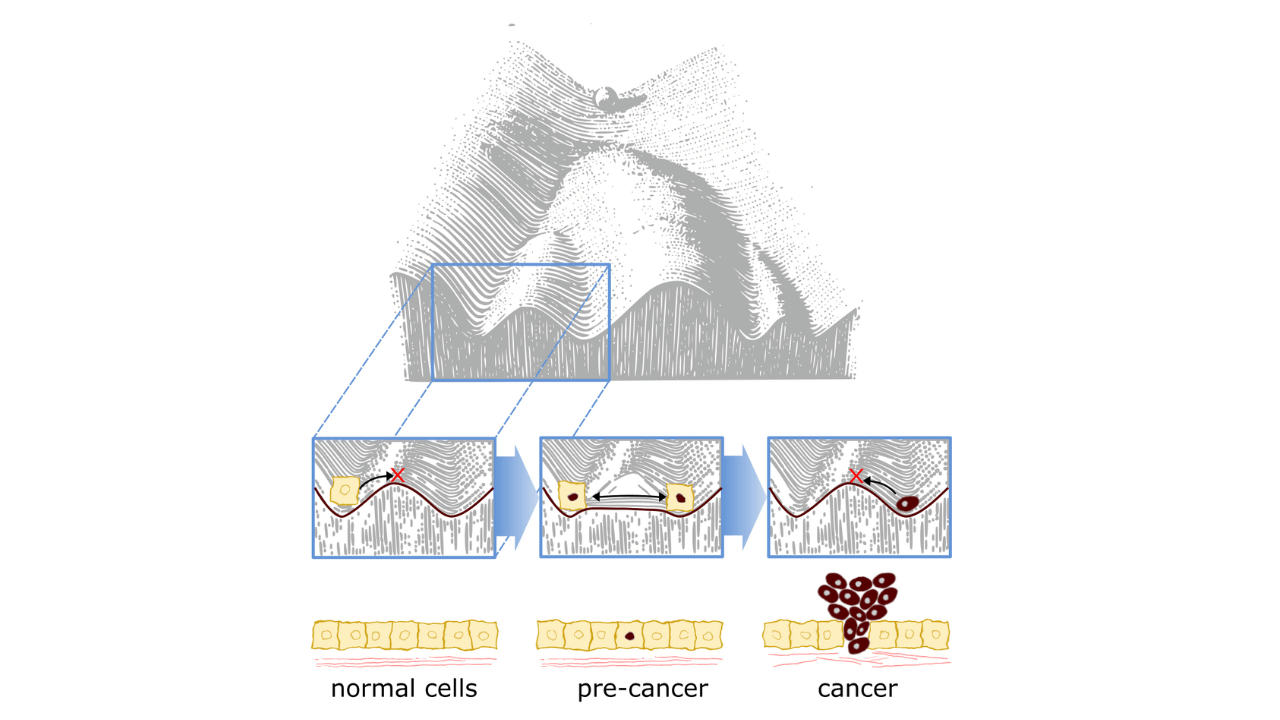
Epigenetics helps define current cell states, yet also shapes how cells respond to external cues such as differentiation or stress. The epigenetic plasticity of a cell describes how flexible this regulation is. Early embryonic cells are highly plastic in that they have the ability to generate all adult cell types. As development progresses, this plasticity is lost as normal healthy adult cells are locked in their identity. Crucially, aberrant reactivation may contribute to pathologies such as cancer. Importantly, we lack a comprehensive molecular understanding of epigenetic plasticity and how it is regulated. This knowledge will be invaluable in opening new avenues for cancer prognostic and therapeutic interventions.
It is increasingly apparent that epigenetic aberrations contribute to all aspects of cancer biology. Cancers often display features of heightened epigenetic plasticity, although what this entails precisely at the molecular level is still being untangled. Hyper-plasticity would confer cancer cells with adaptive advantages as they are readily able adapt to changing environments or therapies, promoting metastasis and relapse. For example, epigenetic hyper-plasticity may underly lineage switching, where cancers reappear after treatment as genetically identical yet functionally distinct pathologies, or acquired drug resistance. It is unknown the extent epigenetic hyper-plasticity occurs in cancers, how this manifests at a molecular level, and the significance this has on cancer progression and response to therapy. Our lab uses insights from developmental epigenetics to understand how the normal tight control of epigenetic plasticity is hijacked by cancers, using single-cell epigenomic, CRISPR and molecular cell biology technologies. Research in the Eckersley-Maslin laboratory is supported by Snow Medical, Peter Macallum Cancer Foundation, The Lorenzo and Pamela Galli Medical Research Trust and The National Stem Cell Foundation of Australia.
Current projects