By targeting oncogenic signalling in cancer and understanding the impact of this therapy on both the tumour cell and its microenvironment, the Grant McArthur lab aims to develop novel treatment strategies that are durable and prevent therapy resistance. The Grant McArthur laboratory has a specific interest in melanoma and pancreatic cancer.
Major themes include:
- Defining the molecular events underpinning therapy response, tolerance and resistance
- Role of cellular metabolism in the response to targeted and immune therapies
- Understanding the impact of the tumour niche on cell plasticity and therapy resistance
- Immunobiology of melanoma
- Translating our preclinical studies into clinical trials
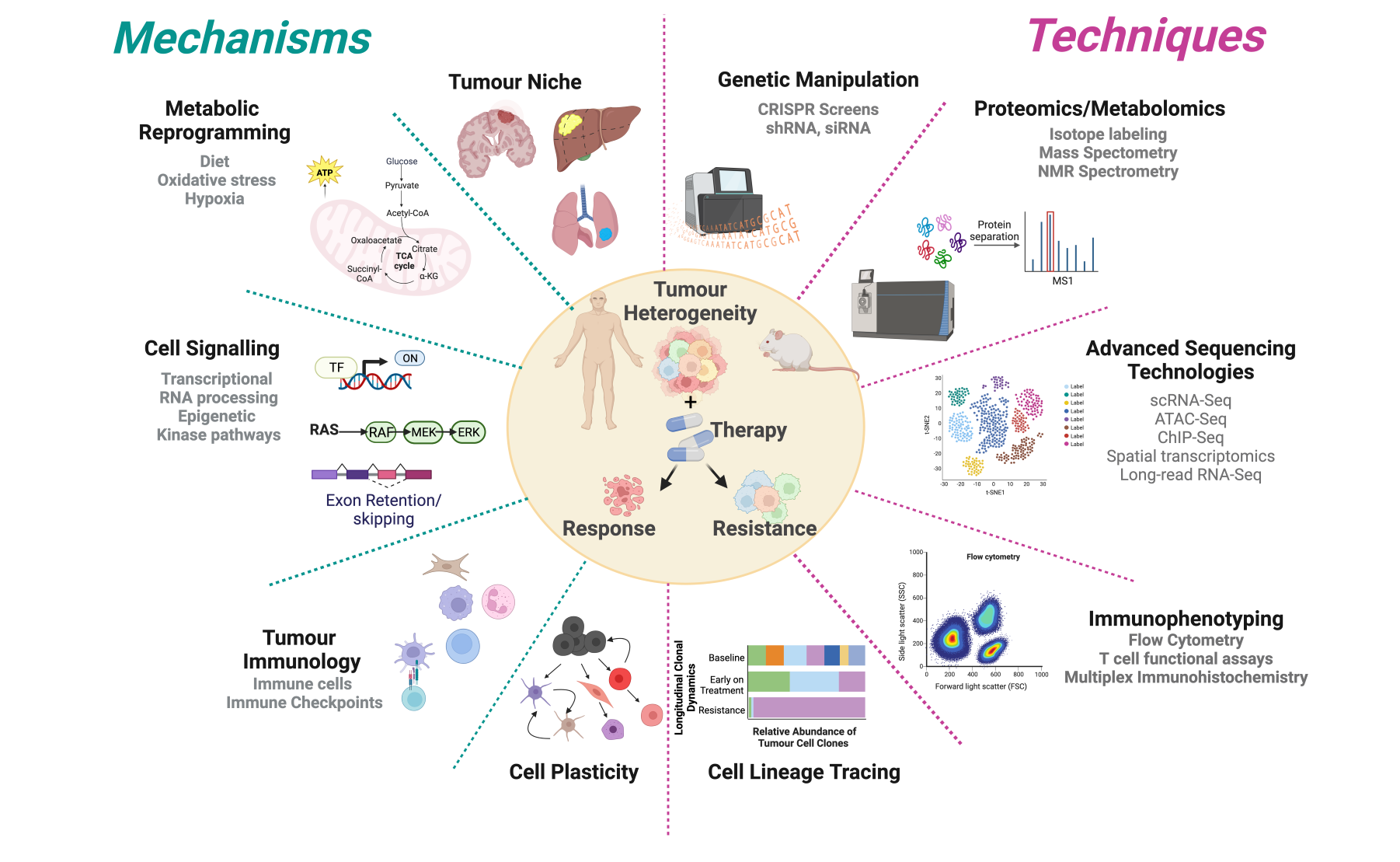
What we study and how
Current projects
Lab members
A/Prof Karen Sheppard; Dr Lorey; Dr Aparna Rao; Dr Reem Saleh; Masaaki Sunaoshi - Visiting Scientist; Darius Schenk - Research Assistant; Riya Patel - PhD student; Dr Michael Lee - PhD student; Dr Veronica Aedo-Lopez - PhD Student; Dr Prachi Bhave - PhD Student; Lydia Lim - PhD student; Shay Arabi - PhD student; Arwa AlKaraki - PhD Student; Dr Roslyn Wallace - Masters student
Related links
Related pages

